Introducing CHIMERYS 5:
Accelerated, Scalable, and Smarter Proteomics Analysis
CHIMERYS 5: Advanced Inference. Expanded PTMs. Absolute Confidence.
-
Spectrum-centric & acquisition-method agnostic (DDA, PRM, DIA) software for proteome research in academia and industry
-
One engine for every instrument - now including timsTOF and multi-vendor raw formats.
-
Enhanced PTM Coverage and Deep-Learning Models: Next-generation AI models delivering unmatched accuracy across an expanded PTM landscape
-
Accelerated search, cloud-native scalability, and seamless integration for enterprise-scale proteomics.
Publication Alert! CHIMERYS paper pubished in Nat. Methods
Unifying the Analysis of Bottom-Up Proteomics Data with CHIMERYS
Frejno, M., Berger, M.T., Tüshaus, J. et al. Unifying the analysis of bottom-up proteomics data with CHIMERYS. Nat Methods 22, 1017–1027 (2025). https://doi.org/10.1038/s41592-025-02663-w
Unifying software for DDA, DIA and PRM data
-
One unifying, spectrum-centric and library-free approach for the analysis of peptide tandem mass spectra irrespective of acquisition type
-
Consistent false discovery rate estimation and post-processing, facilitating fair comparisons of DDA and DIA data
-
Label-free quantification at MS1 or MS2 level, as well as TMT™- and TMTpro™-based quantification
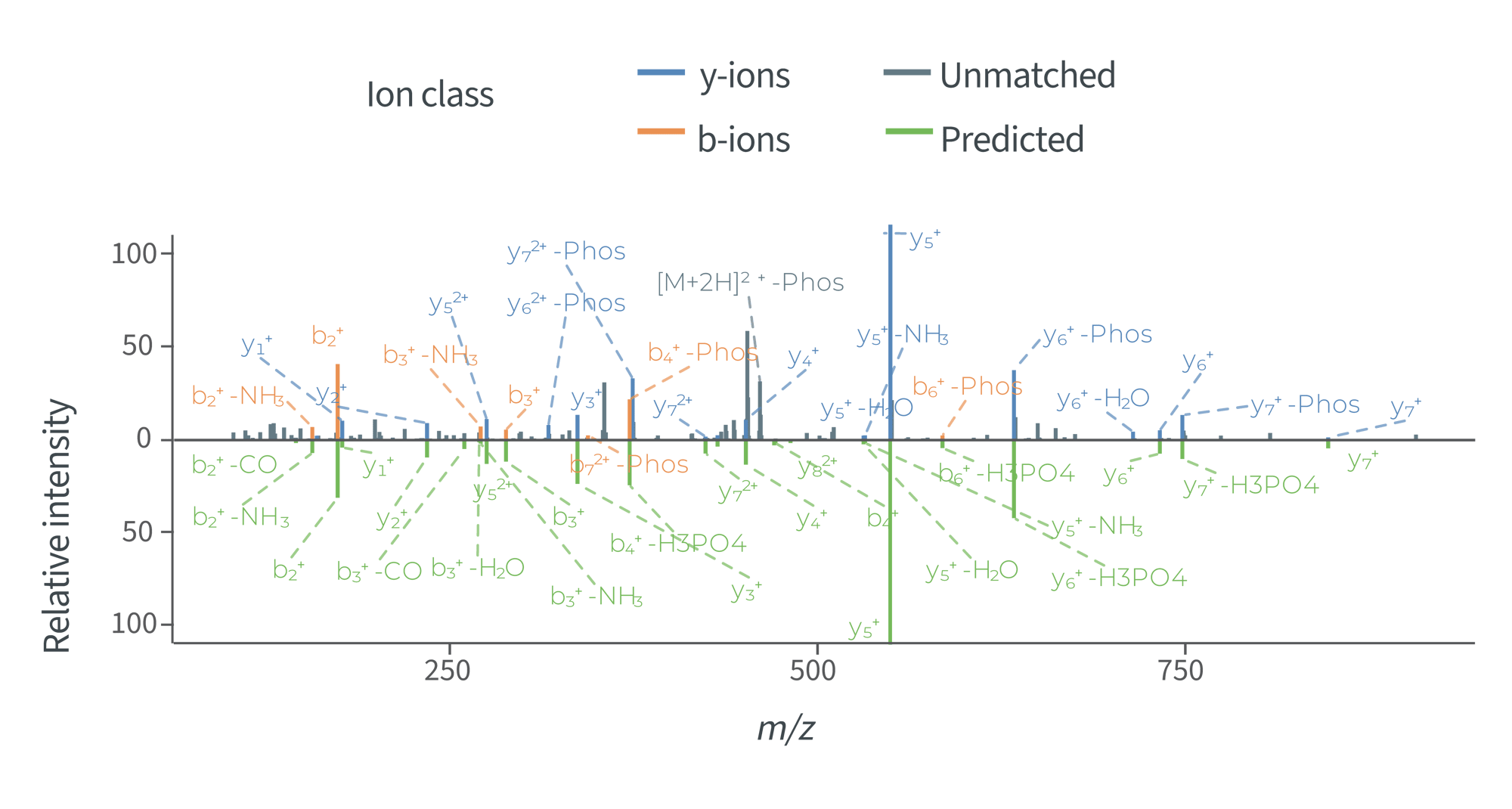

>20 PTMs for your biological R&D
-
Prediction of fragment ion intensities and retention times for over 20 biologically relevant post-translational modifications.
-
Molecular representation using Self-Referencing Embedded Strings (SELFIES) for extrapolation beyond trainign data
-
Novel CHIMERYS-native PTM localization module based on fragment ion intensity predictions
Enhanced User Experience
-
Seamlessly scaling to parallel process large studies with > 1000 raw files on the MSAID cloud platform for proteomics
-
Increased computational efficiency and parallelism: accelerating time to insight for more data on the same subscription plan
-
Improved sensitivity and identification rates to dig even deeper into newly acquired and previously analyzed data

Frequently Asked Questions about CHIMERYS
Explore detailed information on our software's features, practical usage tips, compatibility details, and effective troubleshooting guidance.
Testimonials
"We have been highly impressed with CHIMERYS, which allows us to now quantify >3,000 proteins from single cells. We simply widen our isolation window in a DDA workflow and let CHIMERYS do the work of confidently assigning fragment ions to multiple peptides in each MS2 spectrum.”
"We are using the groundbreaking search algorithm CHIMERYS in our cutting edge single-cell proteomics platform to accurately and sensitively identify large number of peptides and proteins from our samples. CHIMERYS excites us by surpassing conventional search engines by a factor of 2.6 in terms of ID rate.”
Experience CHIMERYS in Proteome Discoverer Software
INFERYS and CHIMERYS are integrated into Thermo Scientific™ Proteome Discoverer™ software. For more information and licensing, please visit thermofisher.com/proteomediscoverer. If you would like to learn about our data analysis services, feel free to contact us!
Thermo Scientific™ Proteome Discoverer™ software, Thermo Scientific™ Orbitrap™, and Thermo Scientific™ Q Exactive™ are trademarks of Thermo Fisher Scientific, Inc. Tandem Mass Tag, TMT™, and TMTpro™ are trademarks of Proteome Sciences plc. SEQUEST is a trademark of the University of Washington.

